Store spectra (NeuroDeveloper format)
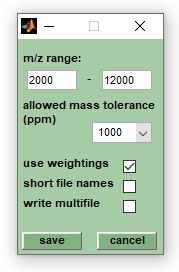
This function allows storing mass spectral data in a format which can be imported by the NeuroDeveloper software. The NeuroDeveloper is a software for teaching and validating artificial neural network models using spectra from various origins (e.g. IR, Raman, or mass spectra). The function export to NeuroDeveloper is available from the File menu bar and can be used for converting peak tables into mass spectra and storing the latter in a simple text format. Select first the spectra to be exported from the listbox denoted with MicrobeMS spectra ID`s (multiple selections are allowed) and chose then export to NeuroDeveloper from the File menu bar. This will open a dialog box entitled export to NeuroDeveloper which allows indicating m/z regions for export (m/z range), chosing the width of the spectral intervals (allowed mass tolerance (ppm)) and selecting the type of intensities (use weightings, or raw MS intensities) to be exported. Please see the screenshot to the right for details.
The extension of the exported NeuroDeveloper data file will be *.txt. Spectra are stored as two-column vectors of m/z values (first column) and the respective MS intensity values (second column). Delimiter is space. All file names are created per default according to the following scheme:
1. class_X - X denotes the spectra's class membership values, aka class labels, which can be defined by
means of the function Class assignment
2. Genus - indicates the genus assignemnt
3. Species - species assignment
4. Strain - strain assignment
5. Y - a sequential number denoting the Y-th spectrum
Further parameters of the dialog box export to NeuroDeveloper:
- use weightings (checkbox): if this checkbox is activated so-called weightings will be stored instead of standard peak intensity values
- short file names (checkbox): if this checkbox has been selected, all file names are formed only by class assignments (see above), by a core file name (to be indicated) and by the number of the Y-th spectrum.
- write multifile (checkbox): all data are stored in a special array format (useful for validation purposes).
In addition to this, a log file is created and stored in the same directory as the NeuroDeveloper spectra files.
Example of a log-file generated by the function export to NeuroDeveloper:
|
log file of the NeuroDeveloper interface of MicrobeMS, date: 15-Jan-2019, time: 18:09:10
Please do not modify this file!
0.0008 - ppm value (defines the spectral resolution)
2000 - mmin, first point in mass spectrum (lower mass, in m/z units)
12000 - mmax, last point in mass spectrum (highest mass, in m/z units)
1 - UseWeightings, barcoded spectrum (0) or not (1)
#- SMO #- BAS #- NRM - Sequence of preprocessing steps
21 - Number of smoothing points (NSP)
100 - Number of intervals for baseline correction (NIV)
1 - Spectra normalized (1) or not (0)
- autocalibration parameters
2000 12000 198.7744 19784.9382 2000 0.1 7 800 1 30 - peak detection parameters
|